Isola, P., Zhu, J. Y., Zhou, T., & Efros, A. A. (2017). Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 1125-1134).
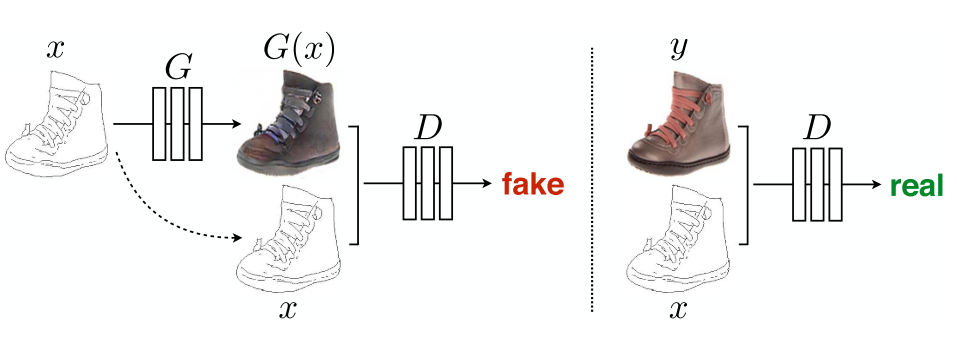
In this paper, the authors address the problem of image to image translation where and image from one domain is translated into another domain. The source image is fed into a ConditionalGenerator network to generate its translated version. Then a SiameseCritic judges if a given pair is real of fake, as shown Fig. 2 of the paper that we copy here:
The Data
We are going to use the CMP Facade Database [1], which is a set of images of building facades and labels indicating different architecural objects, like window, balcony, door, etc. You can read more on details on its web page.
Let's use the untar_data function to download the dataset.There are two files, and we'll use both.
path_base = untar_data(URLs.FACADES_BASE)
path_extended = untar_data(URLs.FACADES_EXTENDED)
get_facade_files = partial(get_image_files, folders=['base', 'extended'])
def get_tuple_files(path):
files = get_facade_files(path)
g_files = groupby(files, lambda x: x.stem).values()
return [sorted(v)[::-1] for v in g_files if v[0].name.split('.')[0]]
files = get_tuple_files(path_base.parent)
ToTensor()(Resize(256)(ImageNTuple.create(files[0]))).show()
We leave cmp_b0068 and cmp_b0331 out of the training set because the authors used those ones as examples in the paper. We are going to do the same.
valid_idx = L(files).itemgot(0).attrgot('stem').argwhere(eq('cmp_b0331'))
valid_idx += L(files).itemgot(0).attrgot('stem').argwhere(eq('cmp_b0068'))
Now we build our datablock with the labels as the xs and the ImageNTuple as the ys. We use 400 images which is what the authors used, as well as random jitter (in this case using fastai presizing).
Very important: we have to use nearest neighbors interpolation, since we have images that are labels.
im_size = 256
facades = DataBlock(
blocks=(ImageTupleBlock, ImageTupleBlock),
get_items=lambda x: get_tuple_files(x)[:402],
get_x=itemgetter(0),
splitter=IndexSplitter(valid_idx),
item_tfms=Resize(286, ResizeMethod.Squish, resamples=(Image.NEAREST, Image.NEAREST)),
batch_tfms=[Normalize.from_stats(0.5*torch.ones(3), 0.5*torch.ones(3)),
*aug_transforms(size=im_size, mult=0.0, max_lighting=0, p_lighting=0, mode='nearest')],
)
dls = facades.dataloaders(path_base.parent, bs=1, num_workers=2)
b = dls.one_batch()
test_eq(len(b), 2)
test_eq(len(b[1]), 2)
test_eq(b[1][0], b[0][0])
dls.show_batch(b, figsize=[5,5])
x = 2*torch.rand(1,6,256,256)-1
cri = Patch70(6)
out = cri(x)
test_eq(out.shape, torch.Size([1,1,30,30]))
show_image(out[0], figsize=[6,6]);float(out.max()),float(out.min())
We can check that the receptive fields are 70x70 as we want
The authors showed that a generator with a UNet structure produced the best results. If we read the paper's section 6.3 Errata we will discover that the original architecture had an extra layer that was not being used. The thinest layer in the UNet used batch normalization, and since it was a 1x1 layer, and the batch size was 1, the normalization was zeroing all the activations. In Pytorch, BatchNorm yields an error in this situation. We skip that layer, as the authors suggest.
We can inspect how many parameters the generator has
g = CGenerator(3, 3, enc_l=5)
print(L(g.parameters()).map(Self.numel()).sum())
We can also check that the output is in the expected range and that it has a normal distribution if the input is normal
g = g.cuda()
x = torch.randn(1, 3, im_size, im_size).cuda()
out = g(x)
test_eq(out.shape, torch.Size([1, 3, im_size, im_size]))
test_eq(out.max()<=1, True)
test_eq(out.max()>=-1, True)
TensorImage(out[0]*0.5+0.5).show()
float(torch.atanh(out).mean()), float(torch.atanh(out).std())
If the input is an image, the output should have some evidence of that
x = b[0]
out = g(x[0])
test_eq(out.shape, torch.Size([1, 3, im_size, im_size]))
test_eq(out.max()<=1, True)
test_eq(out.max()>=-1, True)
TensorImage(out[0]*0.5+0.5).show()
float(torch.atanh(out).mean()), float(torch.atanh(out).std())
We put everything together in a GANLearner.
It is very important to set the value switch_eval=False. If not, the generator will be in eval mode (the BatchNorm layers will behave differently) when the critic is being updated but it will be in trian mode during its SGD step. The same thing would happend for the critic. This leads to model collapse.
We use the optimization hyperparameters that the authors suggest.
Since we are using FixedGANSwitcher we need twice the number of iterations to reproduce the results.
patch70_critic = Patch70(6)
critic = SiameseCritic(patch70_critic)
cgen = CGenerator(3, 3, enc_l=5)
cgen = ConditionalGenerator(cgen)
out = widgets.Output()
learn = GANLearner(
dls, cgen, critic,
GeneratorLoss(GeneratorBCE(), nn.L1Loss(), 1, 100),
crit_bce_loss,
switcher=FixedGANSwitcher(n_crit=1, n_gen=1),
metrics=[l1, GenMetric(gen_bce_loss), CriticMetric(crit_fake_bce), CriticMetric(crit_real_bce)],
opt_func = partial(Adam, mom=0.5, sqr_mom=0.999, wd=0,eps=1e-7),
cbs=[ProgressImage(out, figsize=(20,10), conditional=True)],
gen_first=True,
switch_eval=False)
learn.recorder.train_metrics=True
learn.recorder.valid_metrics=False
out
epochs = 400
learn.fit(epochs, lr=2e-4, wd=0)
learn.show_results(max_n=2, ds_idx=1, figsize=(20,10))
Save the model if you want
We patch the predict method and we can generate output using file names as input.
fn = files[L(files).itemgot(0).attrgot('stem').argwhere(eq('cmp_b0331'))[0]][0]
learn.predict(fn)[0].show();
fn = files[L(files).itemgot(0).attrgot('stem').argwhere(eq('cmp_b0068'))[0]][0]
learn.predict(fn)[0].show();
We put everything together in learner and we can train a pix2pix network with three lines of code. This time we use the DynamicUnet model as generator.
metrics=[l1, GenMetric(gen_bce_loss), CriticMetric(crit_fake_bce), CriticMetric(crit_real_bce)]
out = widgets.Output()
cbs = ProgressImage(out, figsize=(20,10), conditional=True)
learn = pix2pix_learner(dls, resnet34, metrics=metrics, cbs=cbs)
learn.recorder.train_metrics=True
learn.recorder.valid_metrics=False
out
learn.fit(400, lr=2e-4, wd=0)
learn.show_results(max_n=2, ds_idx=1, figsize=(20,10))
